Melb2018
Lab 8: Spacing and distances
This session is concerned with summary statistics for spacings and interpoint distances. The lecturer’s R script is available here (right click and save).
library(spatstat)
## Loading required package: spatstat.data
## Loading required package: methods
## Loading required package: nlme
## Loading required package: rpart
##
## spatstat 1.56-1.007 (nickname: 'Damn You Autocorrect')
## For an introduction to spatstat, type 'beginner'
Exercise 1
For the swedishpines data:
-
Calculate the estimate of the nearest neighbour distance distribution function (G) using
Gest.G <- Gest(swedishpines) -
Plot the estimate of (G(r)) against (r)
plot(G, cbind(km, rs, han) ~ r, main = "Nearest neighbor distance distribution")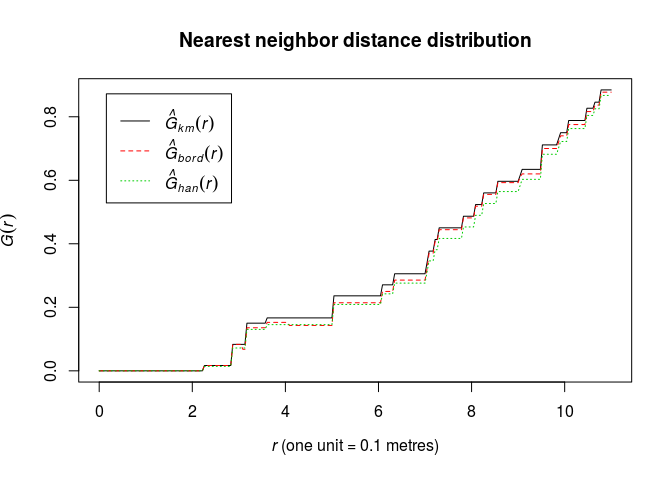
-
Plot the estimate of (G(r)) against the theoretical (Poisson) value (G_{\mbox{pois}}(r) = 1 - \exp(-\lambda \pi r^2)).
E.g.
plot(G, . ~ theo, main = "Nearest neighbor distribution")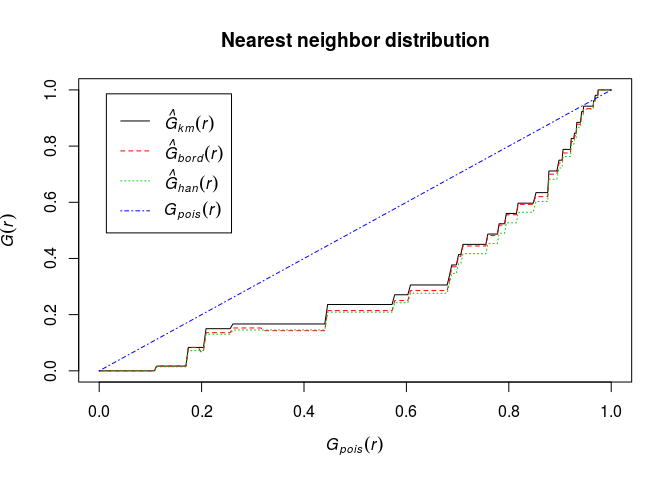
-
Define Fisher’s variance-stabilising transformation for c.d.f.’s by
Phi <- function(x) asin(sqrt(x))Plot the (G) function using the formula
Phi(.) ~ Phi(theo)and interpret it.Phi <- function(x) asin(sqrt(x)) plot(G, Phi(.) ~ Phi(theo), main = "Nearest neighbor distribution")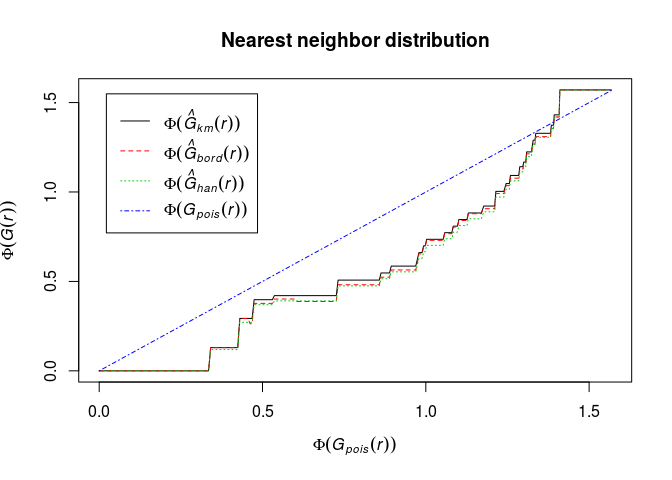
The transformation has made the deviations from CSR in the central part of the curve smaller. We need envelopes to say anything about significance.
Exercise 2
For the swedishpines data:
-
Calculate the estimate of the nearest neighbour distance distribution function (F) using
Fest.Fhat <- Fest(swedishpines) -
Plot the estimate of (F(r)) against (r)
plot(Fhat, main = "Empty Space function")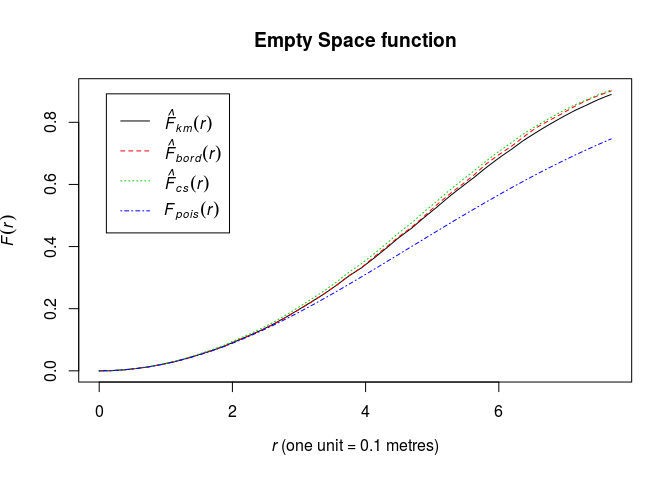
-
Plot the estimate of (F(r)) against the theoretical (Poisson) value (F_{\mbox{pois}}(r) = 1 - \exp(-\lambda \pi r^2)).
plot(Fhat, . ~ theo, main = "")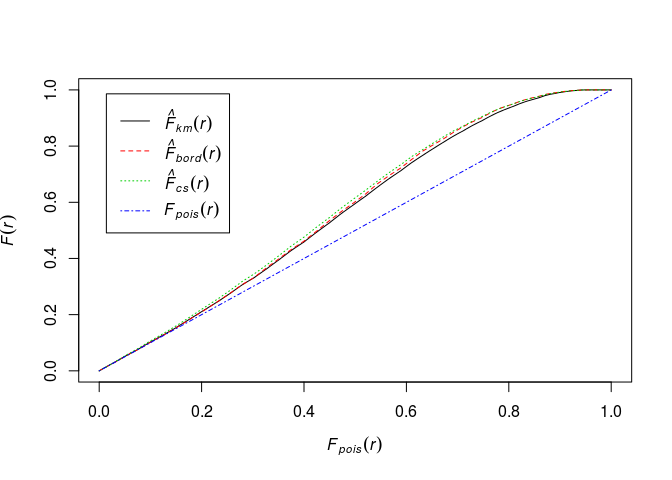
-
Define Fisher’s variance-stabilising transformation for c.d.f.’s by
Phi <- function(x) asin(sqrt(x))Plot the (F) function using the formula
Phi(.) ~ Phi(theo)and interpret it.Phi <- function(x) asin(sqrt(x)) plot(Fhat, Phi(.) ~ Phi(theo), main = "")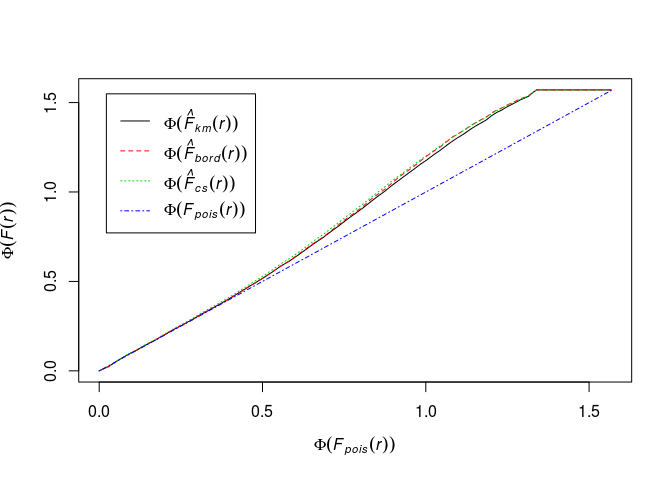
The transformation has changed the picture much. It looks like deviation from CSR, but again, without envelopes it’s hard to draw conclusions.