Melb2018
Lab 3: Intensity dependent on covariate
This session covers tools for investigating intensity depending on a covariate. The lecturer’s R script is available here (right click and save).
library(spatstat)
Exercise 1
The bei dataset gives the locations of trees in a survey area with additional covariate information in a list bei.extra.
-
Assign the elevation covariate to a variable
elevby typingelev <- bei.extra$elevOK, lets do that:
elev <- bei.extra$elev -
Plot the trees on top of an image of the elevation covariate.
plot(elev, main = "") plot(bei, add = TRUE, cex = 0.3, pch = 16, cols = "white")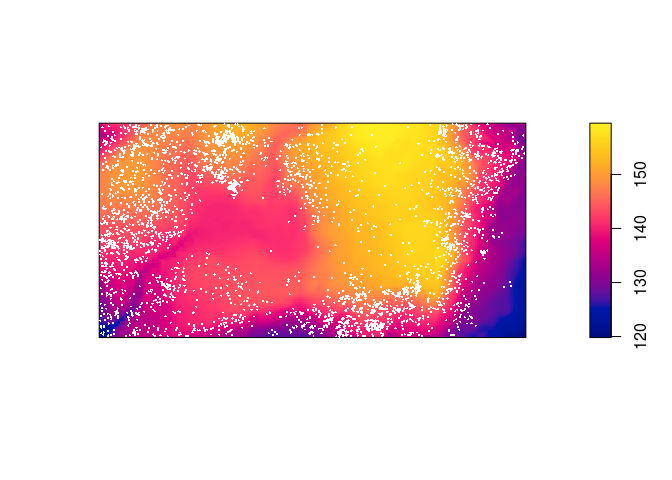
-
Cut the study region into 4 areas according to the value of the terrain elevation, and make a texture plot of the result.
Z <- cut(elev, 4, labels=c("Low", "Med-Low", "Med-High", "High")) textureplot(Z, main = "")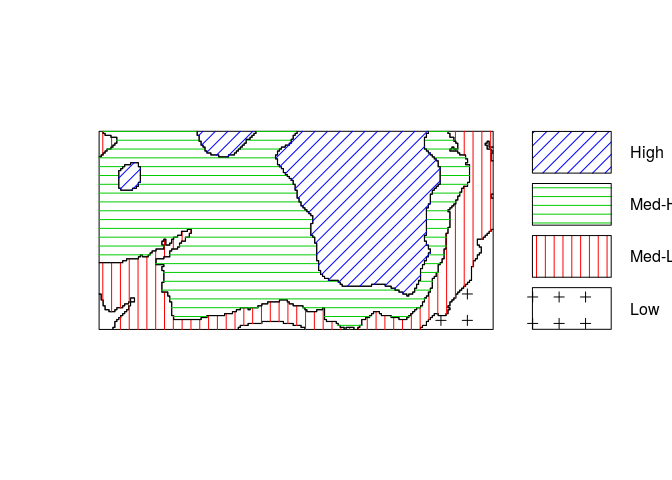
-
Convert the image from above to a tesselation, count the number of points in each region using
quadratcount, and plot the quadrat counts.Y <- tess(image = Z) qc <- quadratcount(bei, tess = Y) -
Estimate the intensity in each of the four regions.
intensity(qc)## tile ## Low Med-Low Med-High High ## 0.002259007 0.006372523 0.008562862 0.005843516
Exercise 2
Assume that the intensity of trees is a function (\lambda(u) = \rho(e(u))) where (e(u)) is the terrain elevation at location u.
-
Compute a nonparametric estimate of the function (\rho) and plot it by
rh <- rhohat(bei, elev) plot(rh)Repeating the R code:
rh <- rhohat(bei, elev) plot(rh)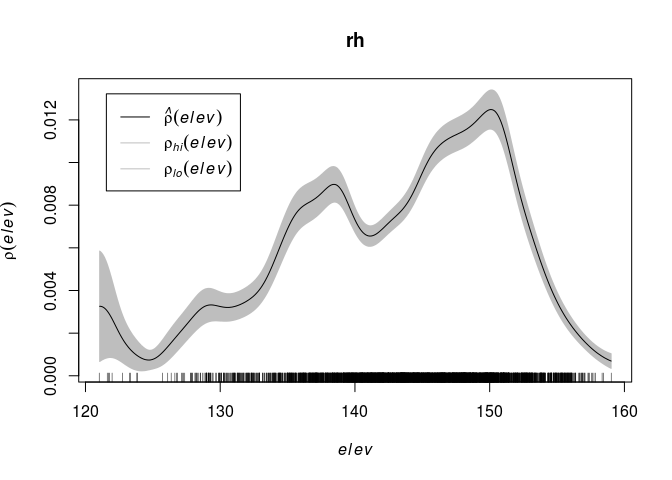
-
Compute the predicted intensity based on this estimate of (\rho).
prh <- predict(rh) plot(prh, main = "") plot(bei, add = TRUE, cols = "white", cex = .2, pch = 16)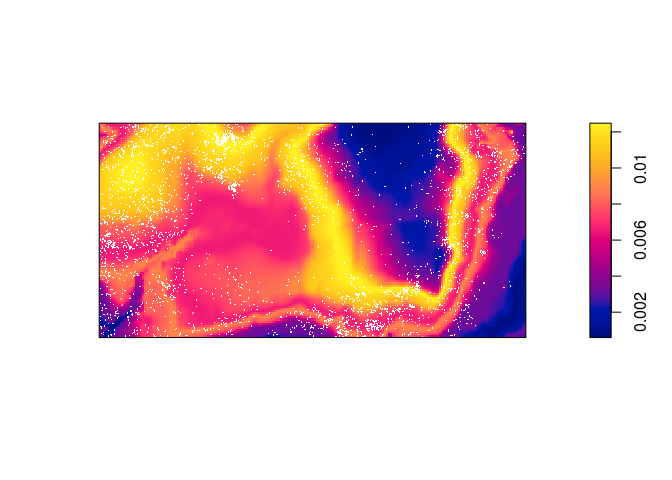
-
Compute a non-parametric estimate by kernel smoothing and compare with the predicted intensity above.
The kernel density estimate of the points is computed and plotted with the following code:
dbei <- density(bei, sigma = bw.scott) plot(dbei, main = "") plot(bei, add = TRUE, cols = "white", cex = .2, pch = 16)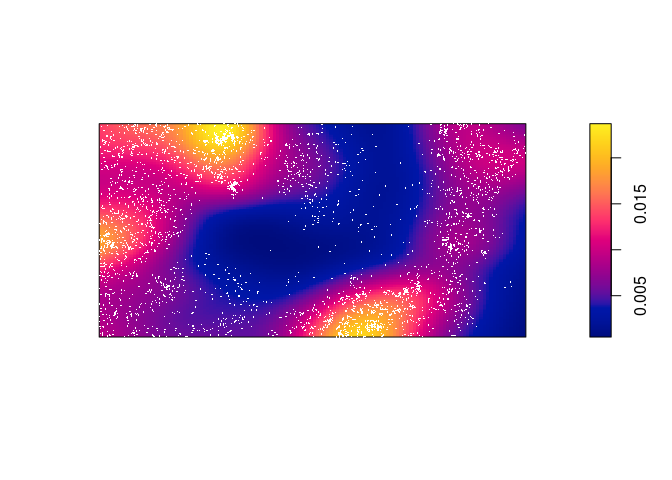
Which seems to be quite different form the predicted intentisty.
-
Bonus info: To plot the two intensity estimates next to each other you collect the estimates as a spatial object list (
solist) and plot the result (the estimates are calledpredandkerbelow):l <- solist(pred, ker) plot(l, equal.ribbon = TRUE, main = "", main.panel = c("rhohat prediction", "kernel smoothing"))l <- solist(prh, dbei) plot(l, equal.ribbon = TRUE, main = "", main.panel = c("rhohat prediction", "kernel smoothing"))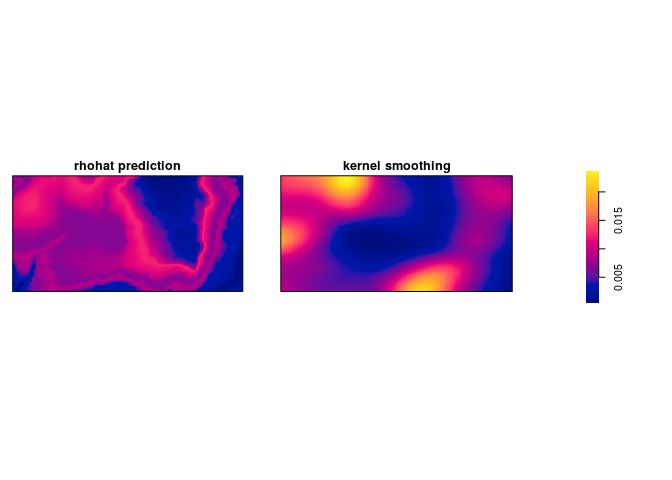
Exercise 3
-
Continuing with the dataset
beiconduct both Berman’s Z1 and Z2 tests for dependence onelev, and plot the results.The tests are done straightforwardly with
berman.test:Z1 <- berman.test(bei, elev) print(Z1)## ## Berman Z1 test of CSR in two dimensions ## ## data: covariate 'elev' evaluated at points of 'bei' ## Z1 = -0.72924, p-value = 0.4659 ## alternative hypothesis: two-sidedplot(Z1)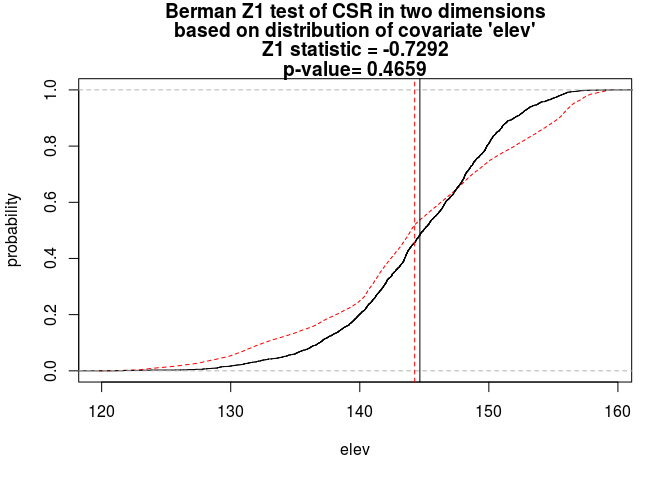
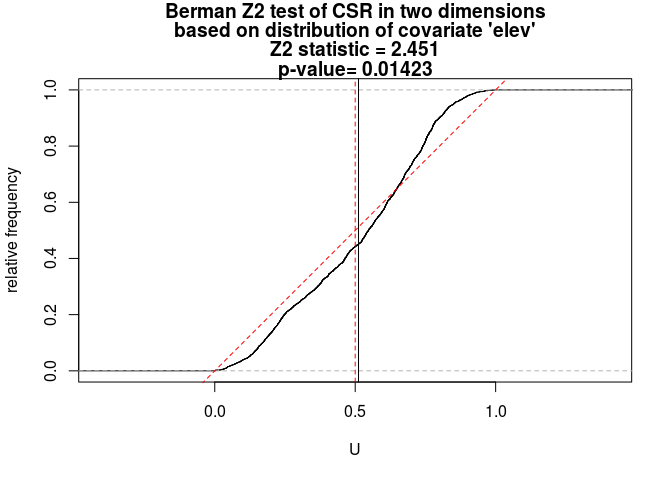
Z2 <- berman.test(bei, elev, which = "Z2") print(Z2)## ## Berman Z2 test of CSR in two dimensions ## ## data: covariate 'elev' evaluated at points of 'bei' ## and transformed to uniform distribution under CSR ## Z2 = 2.4514, p-value = 0.01423 ## alternative hypothesis: two-sidedplot(Z2)