Melb2018
Lab 2: Intensity
This session covers exploratory tools for investigating intensity. The lecturer’s R script is available here (right click and save).
library(spatstat)
Exercise 1
The dataset japanesepines contains the locations of Japanese Black Pine trees in a study region.
-
Plot the
japanesepinesdata.plot(japanesepines)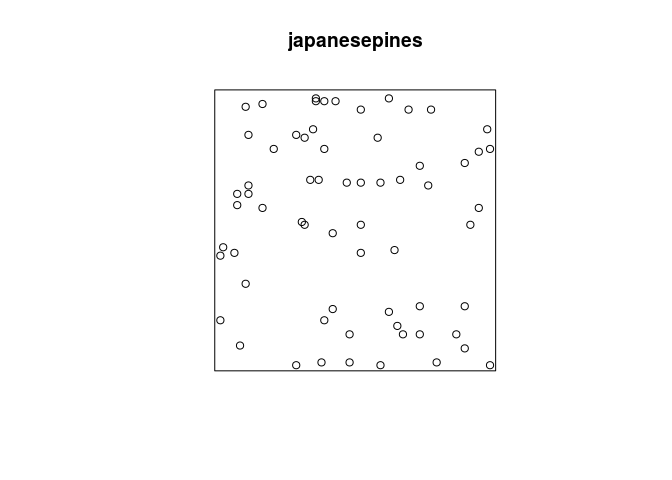
-
Use the command
quadratcountto divide the study region of the Japanese Pines data into a 3x3 array of equal quadrats, and count the number of trees in each quadrat.We count the number of Japanese Pines and print the results:
qc <- quadratcount(japanesepines, nx = 3) qc## x ## y [0,0.333) [0.333,0.667) [0.667,1] ## [0.667,1] 6 15 7 ## [0.333,0.667) 10 4 3 ## [0,0.333) 4 8 8By default,
quadratcountuses the same number of division of the y-axis as given bynx. -
Most plotting commands will accept the argument
add=TRUEand interpret it to mean that the plot should be drawn over the existing display, without clearing the screen beforehand. Use this to plot the Japanese Pines data, and superimposed on this, the 3x3 array of quadrats, with the quadrat counts also displayed.We do the superimposed plotting in the following manner:
plot(qc, main = "") plot(japanesepines, add = TRUE, pch = 16, cex = 0.5)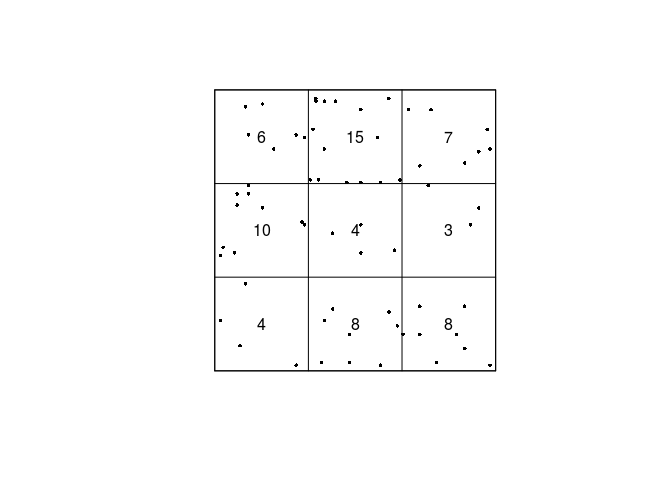
-
Use the command
quadrat.testto perform the (\chi)-square test of CSR on the Japanese Pines data.We do the Chi-squarred test with the following line.
chisq.res <- quadrat.test(qc) print(chisq.res)## ## Chi-squared test of CSR using quadrat counts ## Pearson X2 statistic ## ## data: ## X2 = 15.169, df = 8, p-value = 0.1119 ## alternative hypothesis: two.sided ## ## Quadrats: 3 by 3 grid of tilesAs seen by the P-value, there seems to be no strong evidence for an over- or under-representation of points in any of the quadrats.
-
Plot the Japanese Pines data, and superimposed on this, the 3x3 array of quadrats and the observed, expected and residual counts. Use the argument
cexto make the numerals larger andcolto display them in another colour.To plot the expected, observed, and residual counts we do the following:
plot(chisq.res, main = "", cex = 1.5, col = "red") plot(japanesepines, add = TRUE, cex = 0.5, pch = 16)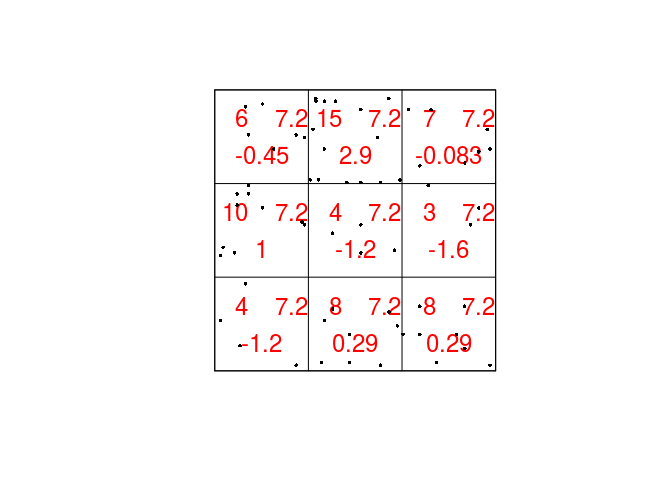
Exercise 2
Japanese Pines, continued:
-
Using
density.ppp, compute a kernel estimate of the spatially-varying intensity function for the Japanese pines data, using a Gaussian kernel with standard deviation (\sigma=0.1) units, and store the estimated intensity in an objectDsay.From the documentation (
?density.ppp) we see that the following will work:D <- density(japanesepines, sigma = 0.1) -
Plot a colour image of the kernel estimate
D.The plotting of the colour image is automatically done by dispatched call to the
plot.immethod by callingploton theimobject.plot(D, main = "")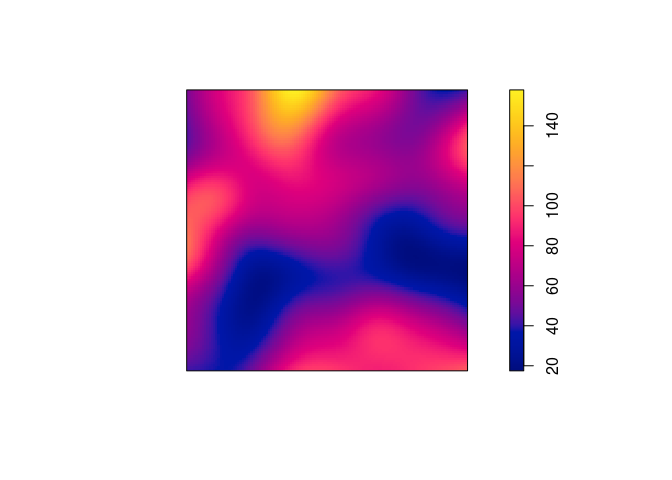
-
Plot a colour image of the kernel estimate
Dwith the original Japanese Pines data superimposed.Again, we can use the
add = TRUEfunctionality of the plotting methods.plot(D, main = "") plot(japanesepines, add = TRUE, cols = "white", cex = 0.5, pch = 16)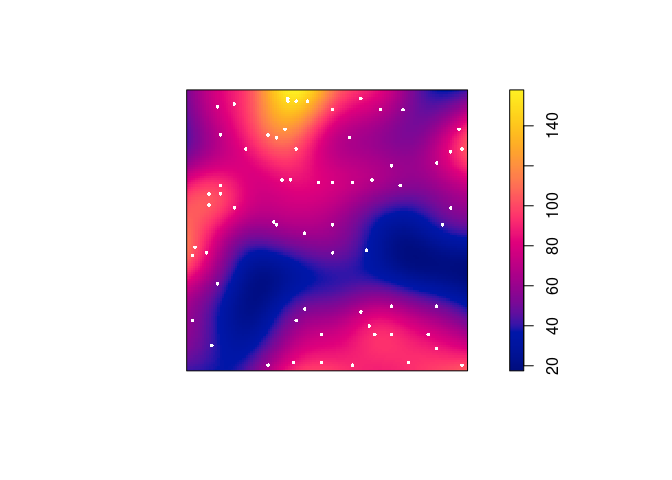
-
Plot the kernel estimate without the ‘colour ribbon’.
From
help("plot.im")we see thatribbon = FALSEdisables the colour key:plot(D, main = "", ribbon = FALSE) plot(japanesepines, add = TRUE, cols = "white", cex = 0.5, pch = 16)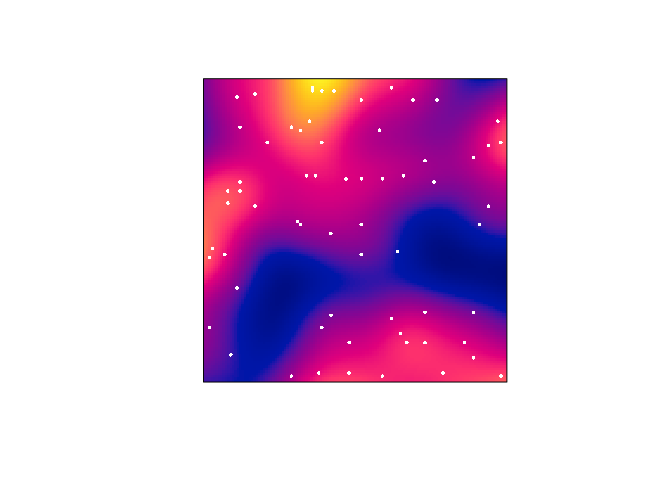
-
Try the following command
persp(D, theta=70, phi=25, shade=0.4)and find the documentation for the arguments
theta,phiandshade.It dispatches to
persp.im, but these arguments are then passed down topersp.defaultthrough the dots (...). From the documentation ofpersp.defaultthey are “angles defining the viewing direction.thetagives the azimuthal direction andphithe colatitude.” Theshadecontrols the shading of the surface facets.persp(D, theta=70, phi=25, shade=0.4, main = "")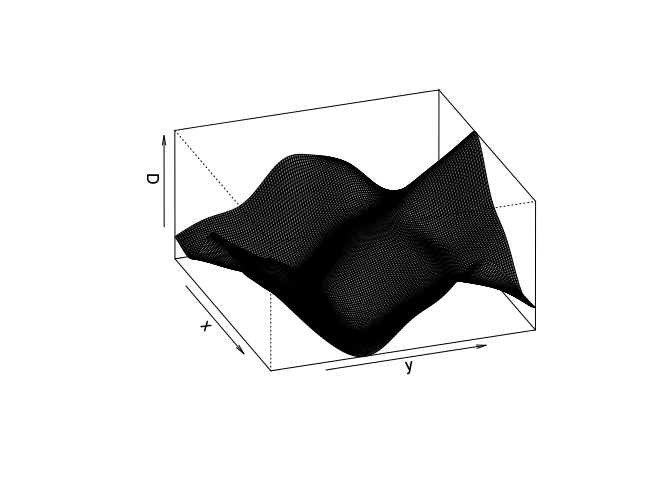
Exercise 3
More Japanese Pines:
-
Compute a kernel estimate of the intensity for the Japanese Pines data using a Gaussian kernel with standard deviation (\sigma=0.15).
As before:
D2 <- density(japanesepines, sigma = 0.15) -
Find the maximum and minimum values of the intensity estimate over the study region. (Hint: Use
summaryorrange)Both
summaryandrangeshow the intensity range:range(D2)## [1] 31.30041 118.22184summary(D2)## real-valued pixel image ## 128 x 128 pixel array (ny, nx) ## enclosing rectangle: [0, 1] x [0, 1] units (one unit = 5.7 metres) ## dimensions of each pixel: 0.00781 x 0.0078125 units ## (one unit = 5.7 metres) ## Image is defined on the full rectangular grid ## Frame area = 1 square units ## Pixel values ## range = [31.30041, 118.2218] ## integral = 63.41479 ## mean = 63.41479 -
The kernel estimate of intensity is defined so that its integral over the entire study region is equal to the number of points in the data pattern, ignoring edge effects. Check whether this is approximately true in this example. (Hint: use
integral)This seems to be true by the following output:
integral(D2)## [1] 63.41479japanesepines## Planar point pattern: 65 points ## window: rectangle = [0, 1] x [0, 1] units (one unit = 5.7 metres)