ECAS2019
Lab 1: Introduction – solution
If you have not already done so, you’ll need to start R and load the
spatstat package by typing
library(spatstat)
Exercise 1
We will study a dataset that records the locations of Ponderosa Pine
trees (Pinus ponderosa) in a study region in the Klamath National
Forest in northern California. The data are included with spatstat as
the dataset ponderosa.
-
assign the data to a shorter name, like
XorP;To assign the data to
Xwe simply write:X <- ponderosa -
plot the data;
To plot the data we do the following:
plot(X)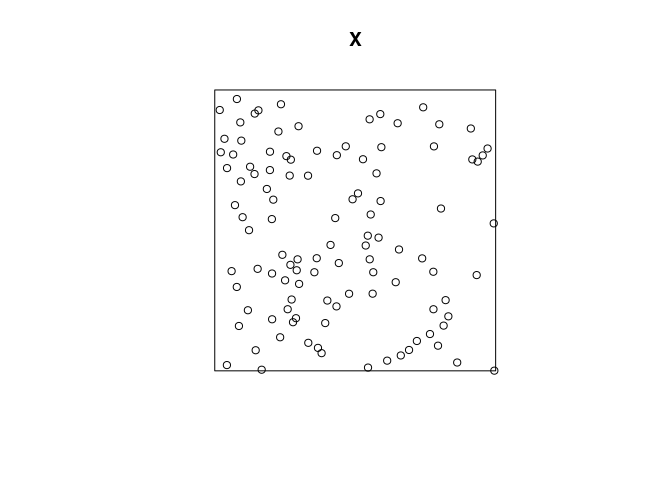
By the S3 method dispatch, this calls the
plot.pppfunction. Thecharsargument indicate that the point type should be be periods (“.”). -
find out how many trees are recorded;
Both
npointsandprint.pppdisplays the number of recorded trees:npoints(X)## [1] 108print(X)## Planar point pattern: 108 points ## window: rectangle = [0, 120] x [0, 120] metresI.e. there are 108 trees in the dataset.
-
find the dimensions of the study region;
The dimensions of the observation window can be seen above. Alternatively, it can be directly assesed via
Window(X)## window: rectangle = [0, 120] x [0, 120] metres -
obtain an estimate of the average intensity of trees (number of trees per unit area).
The average intensity can be computed via
intensity.pppintensity(X)## [1] 0.0075or the more expansive
summary.ppp:summary(X)## Planar point pattern: 108 points ## Average intensity 0.0075 points per square metre ## ## Coordinates are given to 3 decimal places ## i.e. rounded to the nearest multiple of 0.001 metres ## ## Window: rectangle = [0, 120] x [0, 120] metres ## Window area = 14400 square metres ## Unit of length: 1 metre
Exercise 2
The Ponderosa data, continued:
-
When you type
plot(ponderosa), the command that is actually executed isplot.ppp, the plot method for point patterns. Read the help file for the functionplot.ppp, and find out which argument to the function can be used to control the main title for the plot;From the documentation, the argument that controls the title is
mainas is also the case in the regular genericplot. -
plot the Ponderosa data with the title Ponderosa Pine Trees above it;
plot(ponderosa, main = "Ponderosa Pine Trees")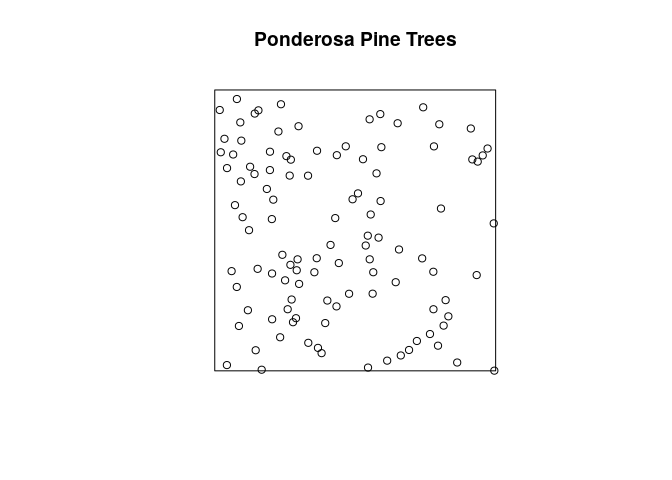
-
from your reading of the help file, predict what will happen if we type
plot(ponderosa, chars="X", cols="green")then check that your guess was correct;
Each point will be plotted as an green “X” and indeed:
plot(ponderosa, chars="X", cols="green")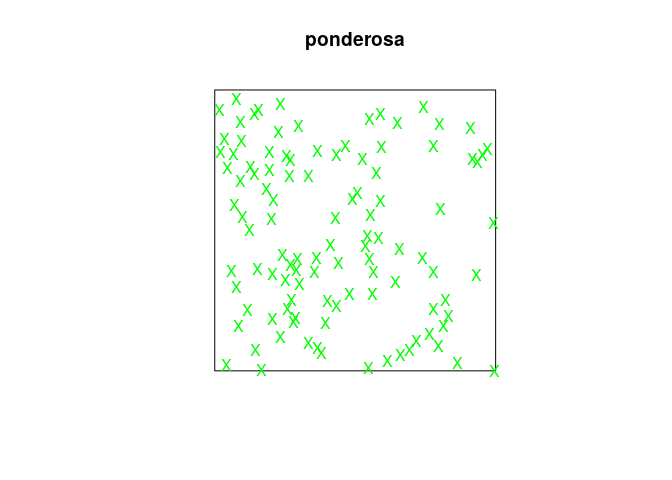
-
try different values of the argument
chars, for example, one of the integers 0 to 25, or a letter of the alphabet. (Note the difference betweenchars=3andchars="+", and the difference betweenchars=4andchars="X").There are subtle differences in the actual character/point types plotted. When given a string literal, the actual character is plotted as the point type.
plot(ponderosa, chars=3, cols="green")
plot(ponderosa, chars="+", cols="green")
plot(ponderosa, chars=4, cols="green")
plot(ponderosa, chars="X", cols="green")
Exercise 3
The dataset hamster is a multitype pattern representing the locations
of cells of two types, dividing and pyknotic.
-
plot the pattern;
plot(hamster, main = "")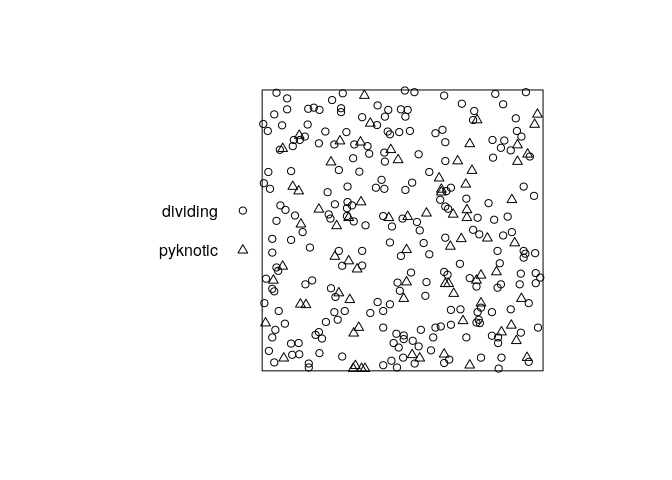
-
plot the pattern again, changing the colours and symbols used to represent the two types of cells;
plot(hamster, main = "", cols = c("red", "blue"), chars = c(3, 5))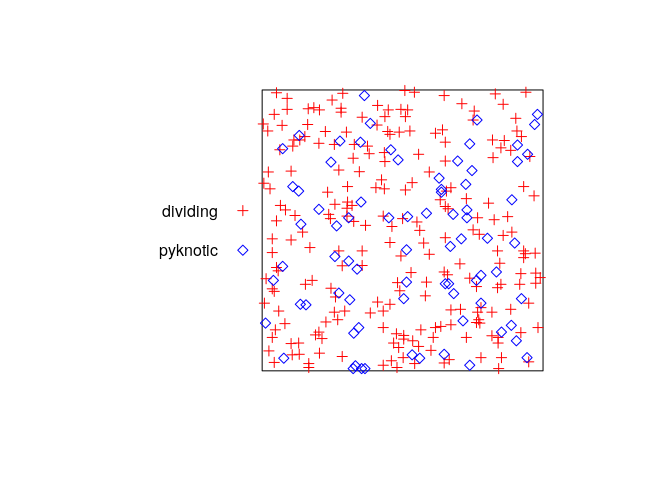
-
plot the patterns of pyknotic and dividing cells separately using
plot(split(hamster)).plot(split(hamster), main = "")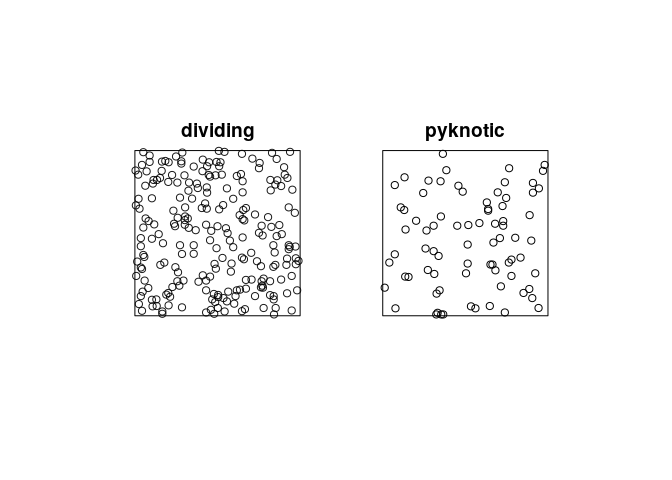
Exercise 4
The following vectors record the locations of 10 scintillation events
observed under a microscope. Coordinates are given in microns, and the
study region was microns, with the origin at the bottom left corner.
x <- c(13, 15, 27, 17, 8, 8, 1, 14, 19, 23)
y <- c(3, 15, 7, 11, 10, 17, 29, 22, 19, 29)
Create a point pattern X from the data, and plot the point pattern
(use owin or square to define the study region).
To define the observation window we do:
w <- owin(c(0, 30), c(0, 30), unitname = c("micron", "microns"))
With this window, we create the ppp object with the funciton of the
same name and plot it:
P <- ppp(x = x, y = y, window = w)
plot(P, main = "Scintillation events")
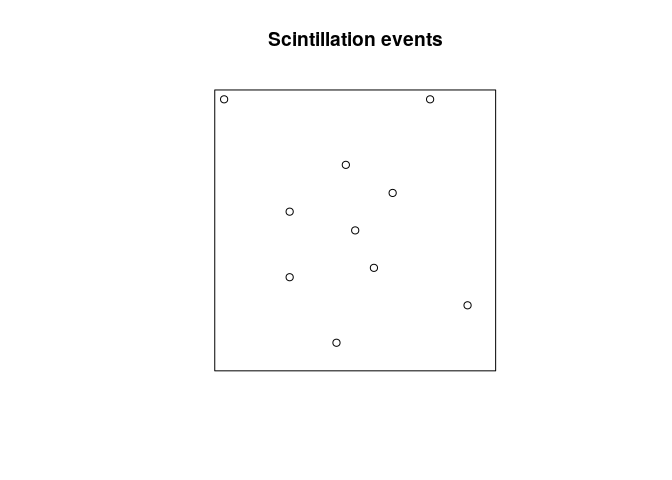
Exercise 5
The file anthills.txt is available in the Data directory on
GitHub and downloadable by this
direct
link
(right click and save).
It records the locations of anthills recorded in a 1200x1500 metre study region in northern Australia. Coordinates are given in metres, along with a letter code recording the ecological ‘status’ of each anthill (in this exercise we will ignore this letter code).
-
read the data into
Ras a data frame, using theRfunctionread.table. (Since the input file has a header line, you will need to use the argumentheader=TRUEwhen you callread.table.)We read the data into R with the line:
dat <- read.table(file = "../Data/anthills.txt", header = TRUE) -
check the data for any peculiarities.
Looking at the first entries of the dataset with
head(dat)## x y status ## 1 105 372 j ## 2 65 142 g ## 3 132 862 f ## 4 22 19 g ## 5 185 1173 e ## 6 69 557 fWe see that there is a
statusvariable indicating that this is a marked point process. -
create a point pattern
hillscontaining these data. Ensure that the unit of length is given its correct name.As before
hills <- with(dat, ppp(x, y, xrange = c(0, 1200), yrange = c(0, 1500), units=c("metre", "metres")))The
pppfunction passes the appropriate arguments toowinwhich we therefore do not need to call explicitly. -
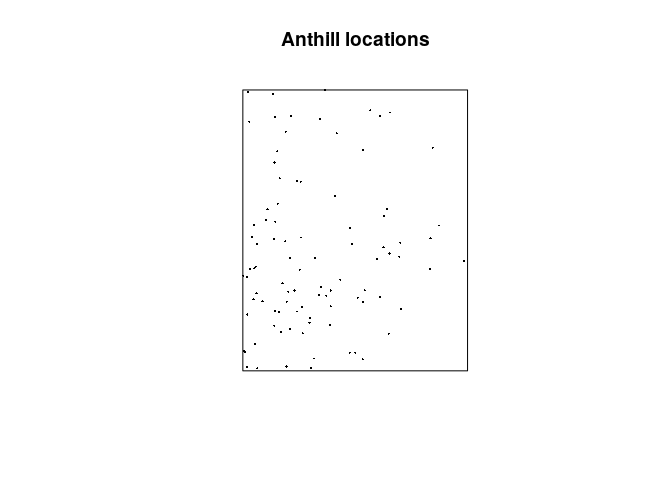
plot the data.
As before, we can now plot the data:
plot(hills, pch = 16, cex = 0.3, main = "Anthill locations")