ECAS2019
Notes for session 2
Adrian Baddeley and Ege Rubak July 15, 2019
Intensity
Often the main objective is to study the “density” of points in the point pattern and to investigate any spatial variation in this density.
Point processes
In a statistical approach to data analysis, we think of the observed data as the outcome of a random process.
To analyse spatial point pattern data, we will regard the observed
point pattern as a
realisation of a random point process
.
It is helpful to visualise a point process as a collection (“ensemble”) of many different possible outcomes. Here is one example:
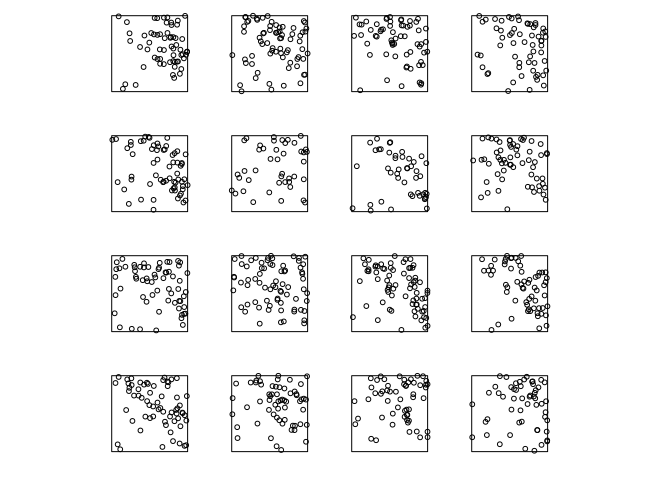
Intensity
The intensity of a point process is the expected number of points per
unit area. It may be a constant , or it may be spatially varying.
Intensity is an average, over all possible outcomes of the point process. We can visualise it by superimposing the ensemble of outcomes:
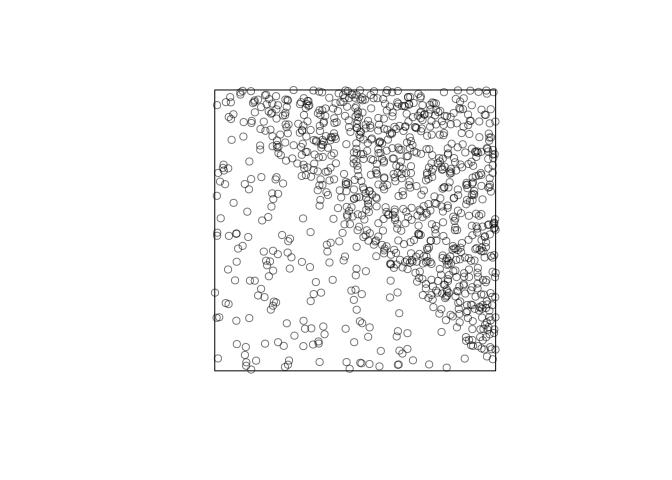
We will usually assume that the point process has an intensity
function
defined at every spatial location
. Then
is the spatially-varying expected number of points per
unit area. It is formally defined to satisfy
for any region , where
denotes the number of points falling in
.
Intensity is closely related to probability density. If
is a point process with
intensity function
, then each individual point inside
has probability density
, where
.
Nonparametric estimation
Because of the close relationship between intensity and probability density, methods for nonparametric estimation of the intensity function are very similar to methods for density estimation.
Nonparametric estimation of spatially-varying intensity
Given a point pattern in a window
the kernel estimate of
intensity is
where is
the smoothing kernel and
is a
correction for edge effects.
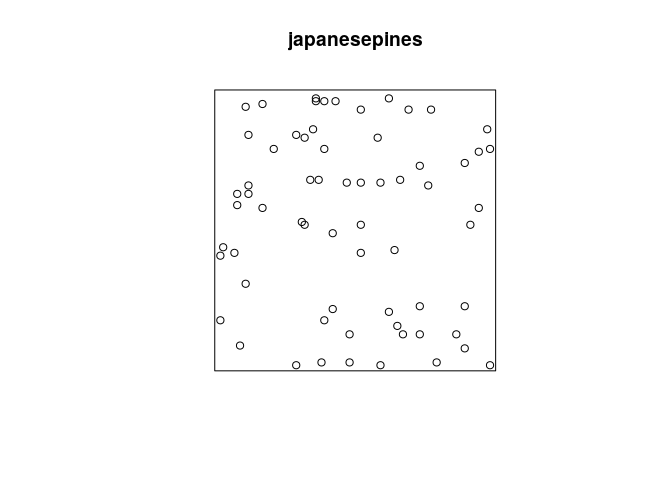
library(spatstat)
plot(japanesepines)
Z <- density(japanesepines, sigma=0.1)
plot(Z)
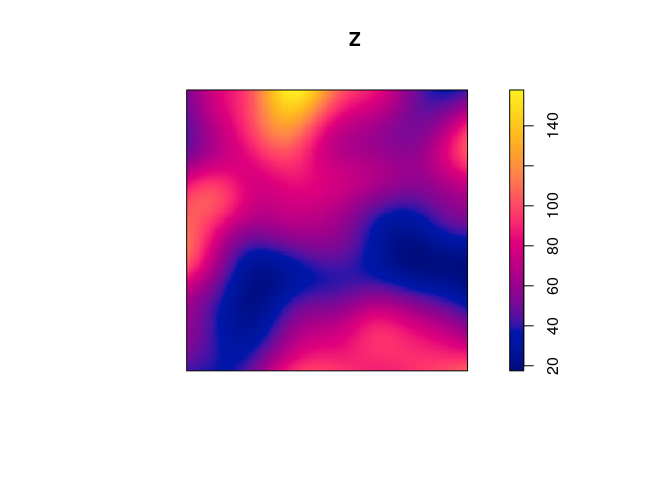
The command in spatstat to compute the kernel estimate of intensity is
density.ppp, a method for the generic function density.
The argument sigma is the bandwidth of the smoothing
kernel.
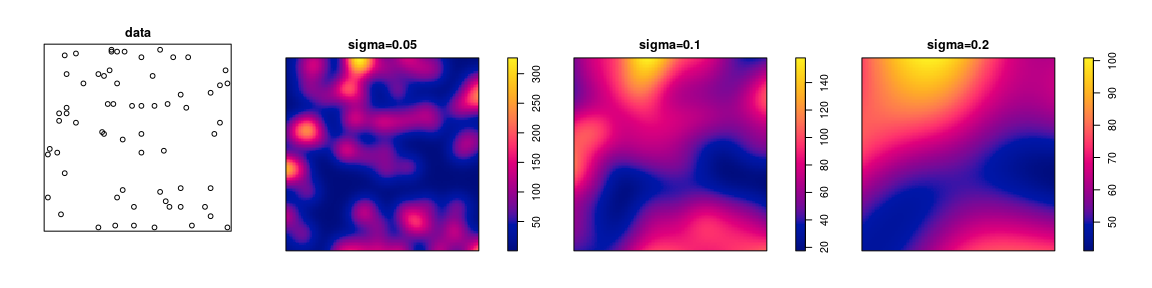
Bandwidth can be selected automatically:
bw.ppl(japanesepines)
## sigma
## 0.7071068
bw.diggle(japanesepines)
## sigma
## 0.05870841
bw.scott(japanesepines)
## sigma.x sigma.y
## 0.1415691 0.1567939
Nonparametric estimation of spatially-varying, mark-dependent intensity
A marked point pattern, with marks which are categorical values, effectively classifies the points into different types.
mucosa
## Marked planar point pattern: 965 points
## Multitype, with levels = ECL, other
## window: rectangle = [0, 1] x [0, 0.81] units
plot(mucosa, cols=c(2,3))
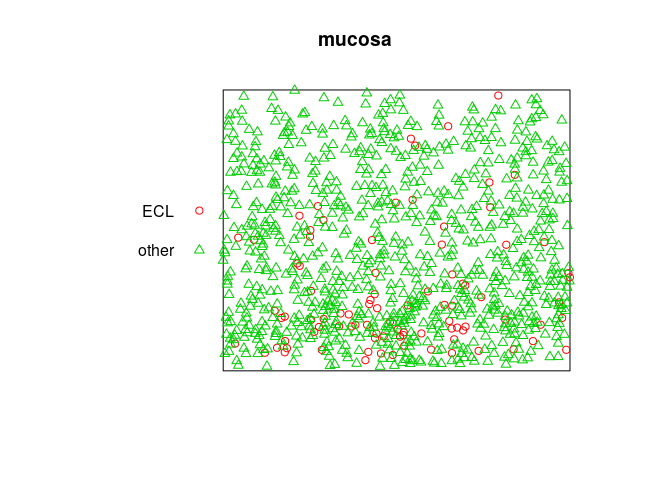
Extract the sub-patterns of points of each type:
M <- split(mucosa)
M
## Point pattern split by factor
##
## ECL:
## Planar point pattern: 89 points
## window: rectangle = [0, 1] x [0, 0.81] units
##
## other:
## Planar point pattern: 876 points
## window: rectangle = [0, 1] x [0, 0.81] units
class(M)
## [1] "splitppp" "ppplist" "solist" "list"
plot(M)
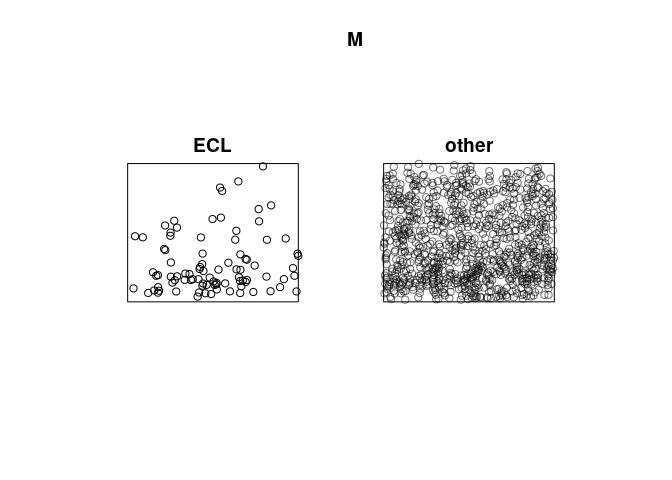
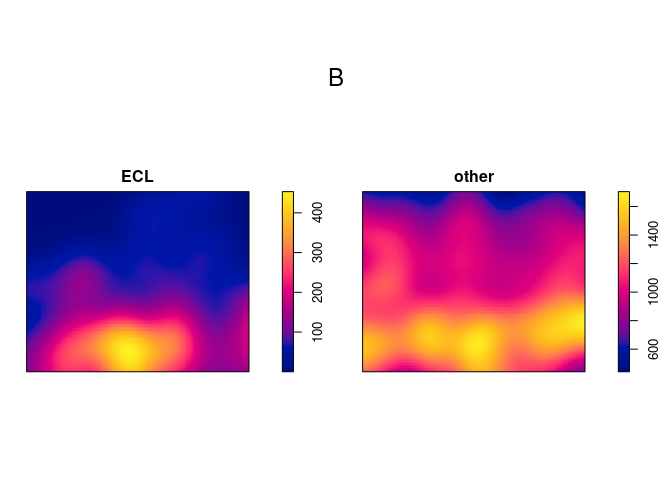
Apply kernel smoothing to each sub-pattern using density.splitppp:
B <- density(M, sigma=bw.ppl)
B
## List of pixel images
##
## ECL:
## real-valued pixel image
## 128 x 128 pixel array (ny, nx)
## enclosing rectangle: [0, 1] x [0, 0.81] units
##
## other:
## real-valued pixel image
## 128 x 128 pixel array (ny, nx)
## enclosing rectangle: [0, 1] x [0, 0.81] units
plot(B)
Suppose
is the intensity function of the points of type
, for
. The intensity function of all points regardless of
type is
Under reasonable assumptions, the probability that a random point at
location belongs to
type
is
We could calculate this by hand in spatstat:
lambdaECL <- B[["ECL"]]
lambdaOther <- B[["other"]]
lambdaDot <- lambdaECL + lambdaOther
pECL <- lambdaECL/lambdaDot
pOther <- lambdaOther/lambdaDot
plot(pECL)
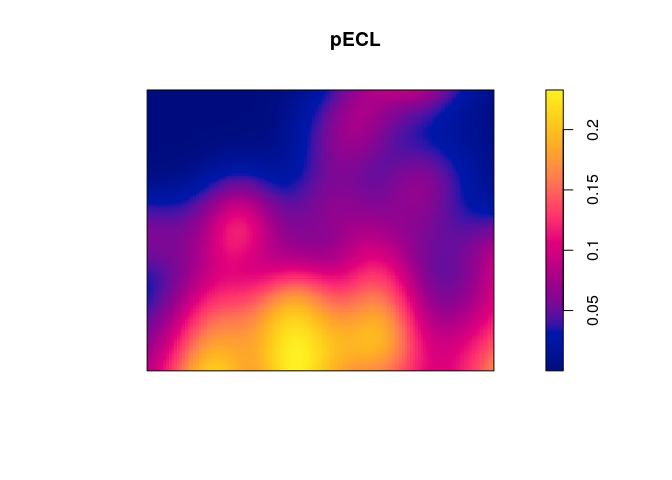
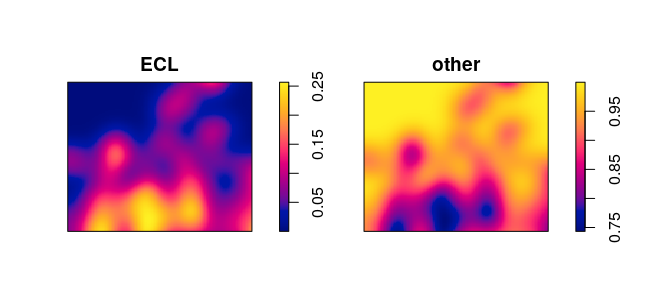
These calculations are automated in the function relrisk (relative
risk):
V <- relrisk(mucosa, bw.ppl, casecontrol=FALSE)
plot(V, main="")
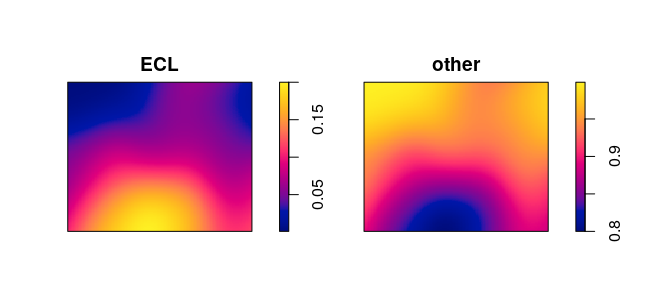
Bandwidth selection for the ratio is different from bandwidth selection
for the intensity. We recommend using the special algorithm
bw.relrisk:
bw.relrisk(mucosa)
## sigma
## 0.1282096
Vr <- relrisk(mucosa, bw.relrisk, casecontrol=FALSE)
plot(Vr, main="")
Segregation of types
“Segregation” occurs if the probability distribution of types of points is spatially varying.
A Monte Carlo test of segregation can be performed using the
nonparametric estimators described above. The function
segregation.test performs it.
segregation.test(mucosa, sigma=0.15, verbose=FALSE)
##
## Monte Carlo test of spatial segregation of types
##
## data: mucosa
## T = 0.33288, p-value = 0.45
Nonparametric estimation of intensity depending on a covariate
In some applications we believe that the intensity depends on a spatial
covariate , in the form
where is an unknown function which we want to estimate. A
nonparametric estimator of
is
where is a
one-dimensional smoothing kernel. This is computed by
rhohat.
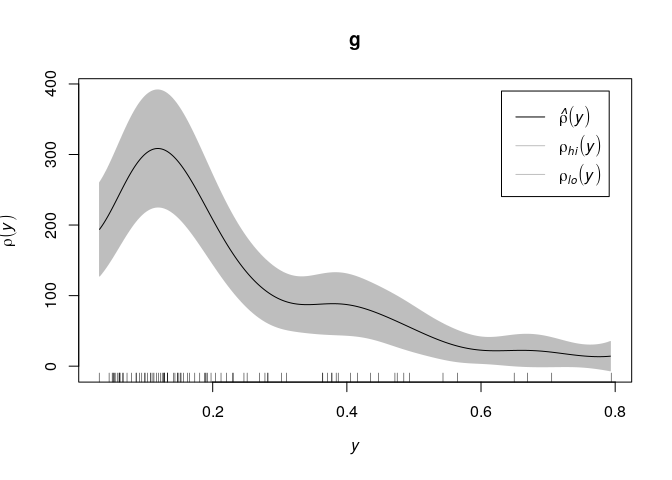
Example: mucosa data, enterochromaffin-like cells (ECL)
E <- split(mucosa)$ECL
plot(E)
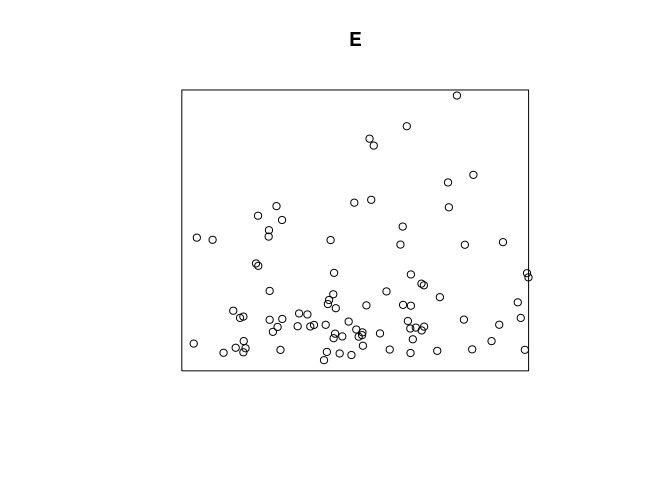
The wall of the gut is at the bottom of the picture. Cell density
appears to decline as we go further away from the wall. Use the string
"y" to refer to the
coordinate:
g <- rhohat(E, "y")
plot(g)
Example: Murchison gold survey.
X <- murchison$gold
L <- murchison$faults
X <- rescale(X, 1000, "km")
L <- rescale(L, 1000, "km")
D <- distfun(L)
plot(solist(gold=X, faults=L, distance=D), main="", equal.scales=TRUE)
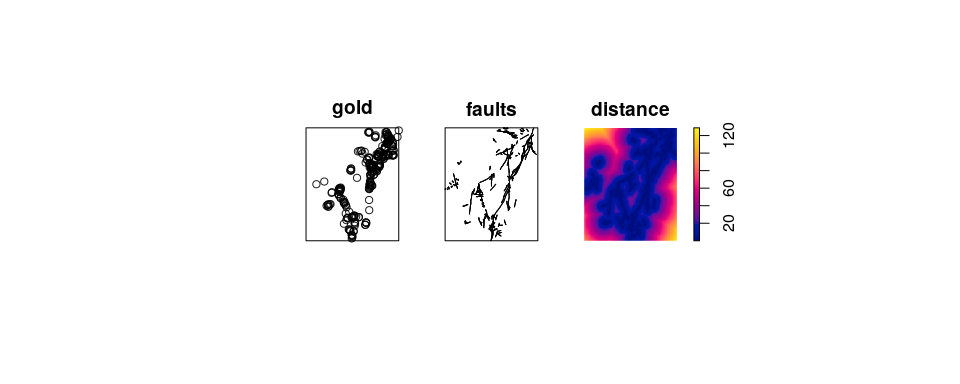
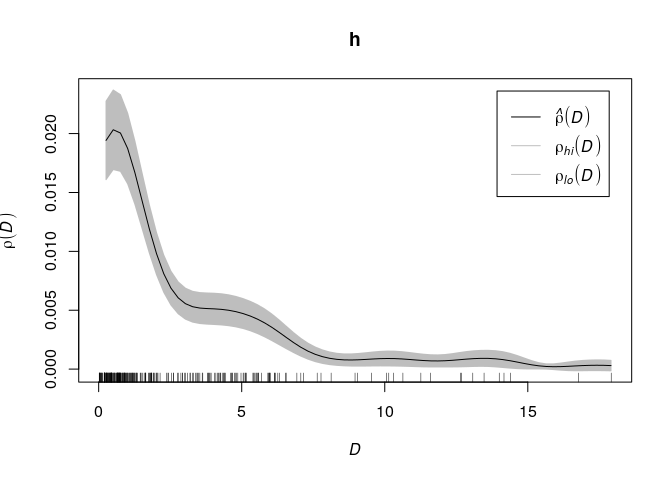
Gold deposits are frequently found near a geological fault line. Here we
converted the fault line pattern into a spatial covariate
h <- rhohat(X, D)
plot(h)
Parametric modelling
We can formulate a parametric model for the intensity and fit it to the
point pattern data, using the spatstat function ppm (point process
model).
In its simplest form, ppm fits a Poisson point process model to the
point pattern data.
Poisson point process
The homogeneous Poisson process with intensity in two-dimensional space is characterised by the
following properties:
- for any region
, the random number
of points falling in
follows a Poisson distribution;
- for any region
, the expected number of points falling in
is
;
- for any region
, given that
, the
points are independent and uniformly distributed inside
;
- for any disjoint regions
, the numbers
of points falling in each region are independent random variables.
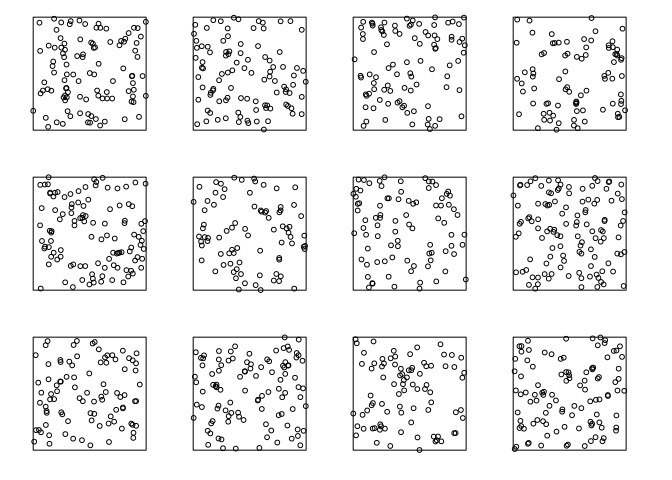
Here are some realisations of the homogeneous Poisson process with intensity 100 (points per unit area):
plot(rpoispp(100, nsim=12), main="", main.panel="")
The inhomogeneous Poisson process with intensity function
is characterised by the following properties:
- for any region
, the random number
of points falling in
follows a Poisson distribution;
- for any region
, the expected number of points falling in
is
- for any region
, given that
, the
points are independent and identically distributed inside
with probability density
, where
;
- for any disjoint regions
, the numbers
of points falling in each region are independent random variables.
Here are some realisations of the inhomogeneous Poisson process with
intensity function :
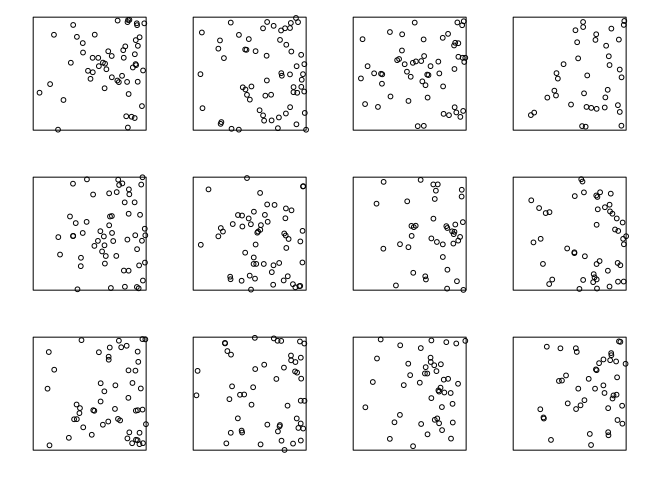
lam <- function(x,y) { 100 * x}
plot(rpoispp(lam, nsim=12), main="", main.panel="")
Loglinear model for intensity
ppm can fit a Poisson point process model to the point pattern data
by maximum likelihood.
A Poisson point process is completely specified by its intensity function. So the procedure for formulating a Poisson model is simply to write a mathematical expression for the intensity function.
In ppm the intensity is assumed to be a loglinear function of the
parameters. That is,
where are parameters to be estimated, and
are spatial covariates.
To fit this model to a point pattern dataset X, we type
ppm(X ~ Z1 + Z2 + .. Zp)
where Z1, Z2, ..., Zp are pixel images or functions.
Important notes:
-
The model is expressed in terms of the log of the intensity.
-
The covariates
(called the “canonical covariates”) can be anything; they are not necessarily the same as the original variables that we were given; they could be transformations and combinations of the original variables.
Fit by maximum likelihood
The Poisson process with intensity function
, controlled by a parameter vector
, has
log-likelihood
The value of which maximises
is taken as the parameter estimate
.
From we can compute the fitted intensity
and hence we can
generate simulated realisations.
Using the likelihood we are able to compute confidence intervals, perform analysis of deviance, conduct hypothesis tests, etc.
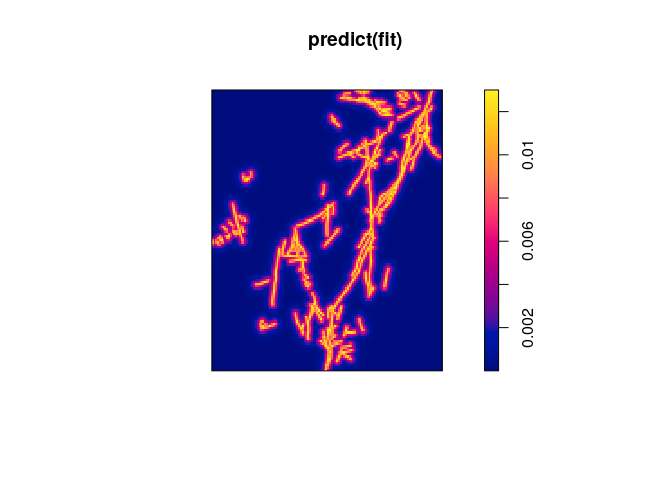
Example: Murchison gold data
Using the Murchison data from above,
fit <- ppm(X ~ D)
The formula implies that the model is
where is
the distance covariate (distance from location
to nearest geological
fault) and
are the regression coefficients. In other words,
the model says that the intensity of gold deposits is an exponentially
decreasing function of distance from the nearest fault.
The result of ppm is a fitted model object of class "ppm". There are
many methods for this class:
fit
## Nonstationary Poisson process
##
## Log intensity: ~D
##
## Fitted trend coefficients:
## (Intercept) D
## -4.3412775 -0.2607664
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) -4.3412775 0.08556260 -4.5089771 -4.1735779 *** -50.73802
## D -0.2607664 0.02018789 -0.3003339 -0.2211988 *** -12.91697
coef(fit)
## (Intercept) D
## -4.3412775 -0.2607664
confint(fit)
## 2.5 % 97.5 %
## (Intercept) -4.5089771 -4.1735779
## D -0.3003339 -0.2211988
anova(fit, test="Chi")
## Analysis of Deviance Table
## Terms added sequentially (first to last)
##
## Df Deviance Npar Pr(>Chi)
## NULL 1
## D 1 590.92 2 < 2.2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
plot(predict(fit))
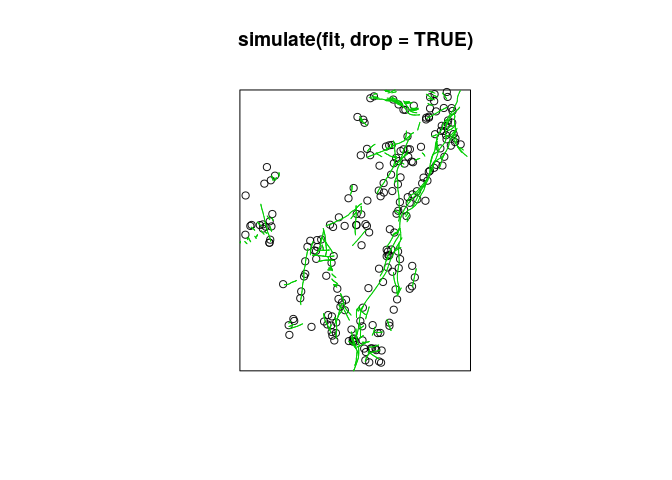
plot(simulate(fit, drop=TRUE))
plot(L, add=TRUE, col=3)
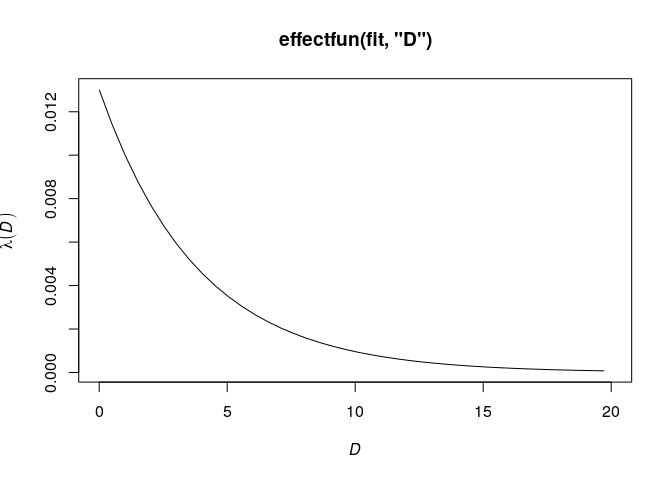
To visualise the intensity of the model as a function of one of the
covariates, we can use the command effectfun:
plot(effectfun(fit, "D"), xlim=c(0, 20))
Example: Japanese Pines data
plot(japanesepines, pch=16)
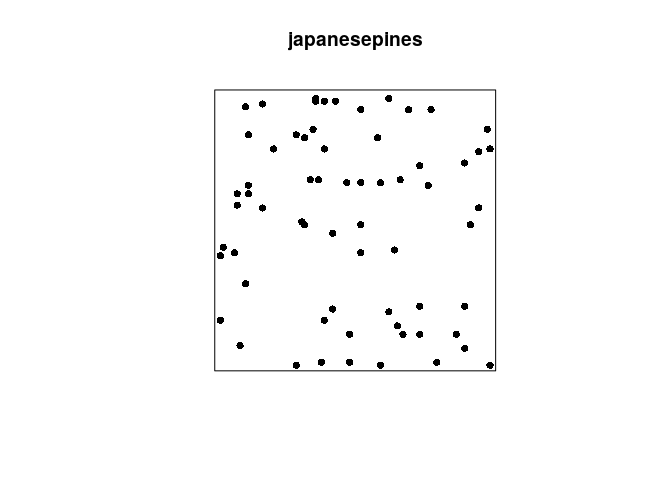
The symbols x, y refer to the Cartesian coordinates, and can be used
to model spatial variation in the intensity when no other covariates are
available:
Jfit <- ppm(japanesepines ~ x + y)
Jfit
## Nonstationary Poisson process
##
## Log intensity: ~x + y
##
## Fitted trend coefficients:
## (Intercept) x y
## 4.0670790 -0.2349641 0.4296171
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 4.0670790 0.3341802 3.4120978 4.7220602 *** 12.1703167
## x -0.2349641 0.4305456 -1.0788181 0.6088898 -0.5457357
## y 0.4296171 0.4318102 -0.4167154 1.2759495 0.9949211
confint(Jfit)
## 2.5 % 97.5 %
## (Intercept) 3.4120978 4.7220602
## x -1.0788181 0.6088898
## y -0.4167154 1.2759495
Jfit2 <- ppm(japanesepines ~ polynom(x,y,2))
Jfit2
## Nonstationary Poisson process
##
## Log intensity: ~x + y + I(x^2) + I(x * y) + I(y^2)
##
## Fitted trend coefficients:
## (Intercept) x y I(x^2) I(x * y) I(y^2)
## 4.0645501 1.1436854 -1.5613621 -0.7490094 -1.2009245 2.5061569
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 4.0645501 0.6670766 2.7571041 5.371996 *** 6.0930788
## x 1.1436854 1.9589569 -2.6957995 4.983170 0.5838237
## y -1.5613621 1.8738722 -5.2340841 2.111360 -0.8332277
## I(x^2) -0.7490094 1.7060242 -4.0927554 2.594737 -0.4390380
## I(x * y) -1.2009245 1.4268186 -3.9974376 1.595589 -0.8416799
## I(y^2) 2.5061569 1.6013679 -0.6324664 5.644780 1.5650101
plot(predict(Jfit2))
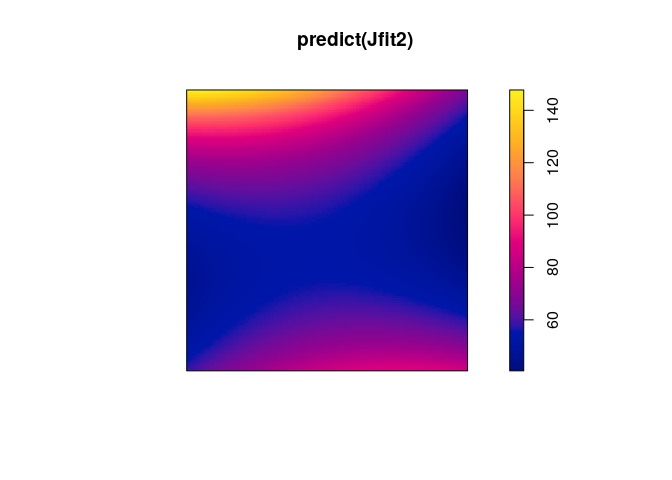
anova(Jfit, Jfit2, test="Chi")
## Analysis of Deviance Table
##
## Model 1: ~x + y Poisson
## Model 2: ~x + y + I(x^2) + I(x * y) + I(y^2) Poisson
## Npar Df Deviance Pr(>Chi)
## 1 3
## 2 6 3 3.3851 0.336
step(Jfit2)
## Start: AIC=-405.35
## ~x + y + I(x^2) + I(x * y) + I(y^2)
##
## Df AIC
## - I(x^2) 1 -407.15
## - x 1 -407.00
## - y 1 -406.67
## - I(x * y) 1 -406.63
## <none> -405.35
## - I(y^2) 1 -404.97
##
## Step: AIC=-407.15
## ~x + y + I(x * y) + I(y^2)
##
## Df AIC
## - x 1 -408.96
## - I(x * y) 1 -408.47
## - y 1 -408.45
## <none> -407.15
## - I(y^2) 1 -406.77
##
## Step: AIC=-408.96
## ~y + I(x * y) + I(y^2)
##
## Df AIC
## - I(x * y) 1 -410.17
## - y 1 -409.78
## <none> -408.96
## - I(y^2) 1 -408.48
##
## Step: AIC=-410.17
## ~y + I(y^2)
##
## Df AIC
## - y 1 -410.51
## <none> -410.17
## - I(y^2) 1 -409.66
##
## Step: AIC=-410.51
## ~I(y^2)
##
## Df AIC
## - I(y^2) 1 -410.67
## <none> -410.51
##
## Step: AIC=-410.67
## ~1
## Stationary Poisson process
## Intensity: 65
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## log(lambda) 4.174387 0.1240347 3.931284 4.417491 *** 33.65499
plot(simulate(Jfit2), main = "")
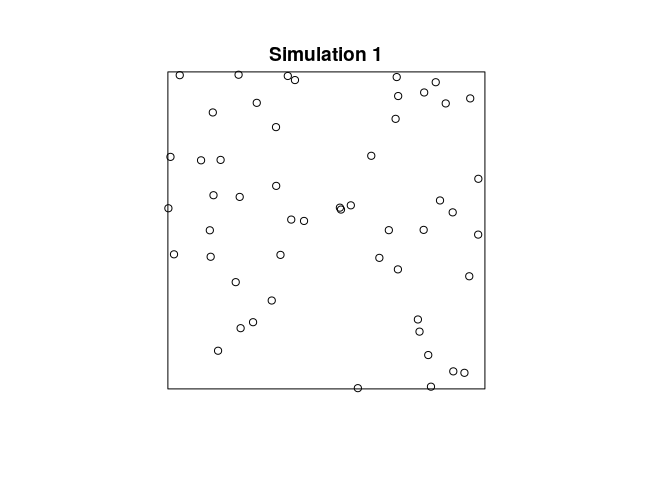
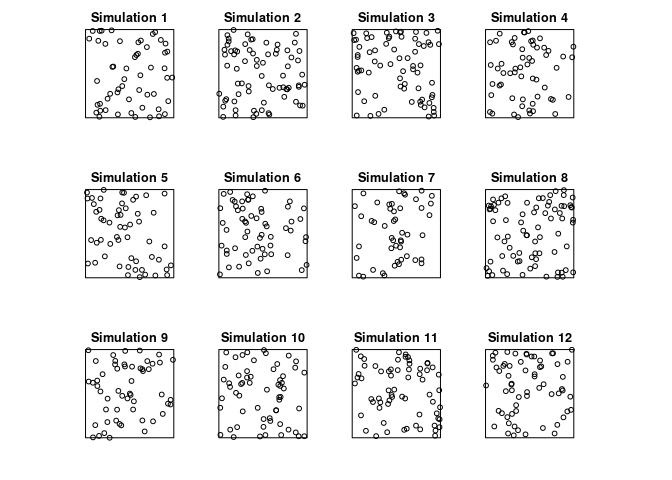
plot(simulate(Jfit2, nsim=12), main = "")
## Generating 12 simulated patterns ...1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12.
Intensity depending on marks
In a multi-type point pattern the points have marks which are categorical values:
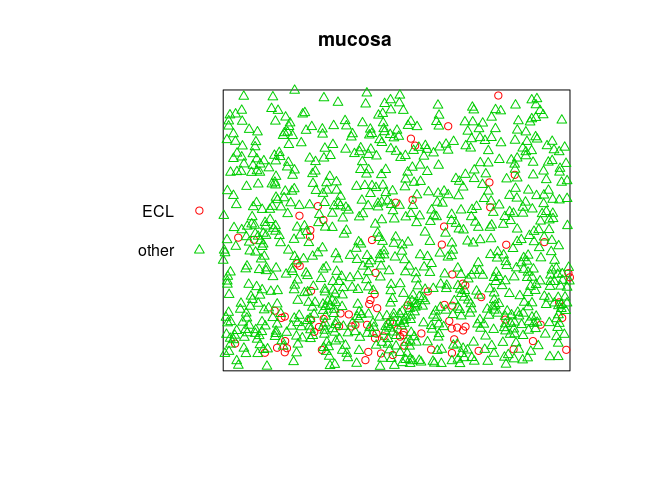
mucosa
## Marked planar point pattern: 965 points
## Multitype, with levels = ECL, other
## window: rectangle = [0, 1] x [0, 0.81] units
plot(mucosa, cols=c(2,3))
We can fit a Poisson model in which the intensity depends on the type of
point, using the variable name marks in the model formula.
model0 <- ppm(mucosa ~ marks)
model0
## Stationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks
##
## Intensities:
## beta_ECL beta_other
## 109.8765 1081.4815
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 4.699357 0.1059998 4.491602 4.907113 *** 44.33365
## marksother 2.286730 0.1112542 2.068675 2.504784 *** 20.55409
coef(model0)
## (Intercept) marksother
## 4.699357 2.286730
plot(predict(model0), equal.ribbon=TRUE)
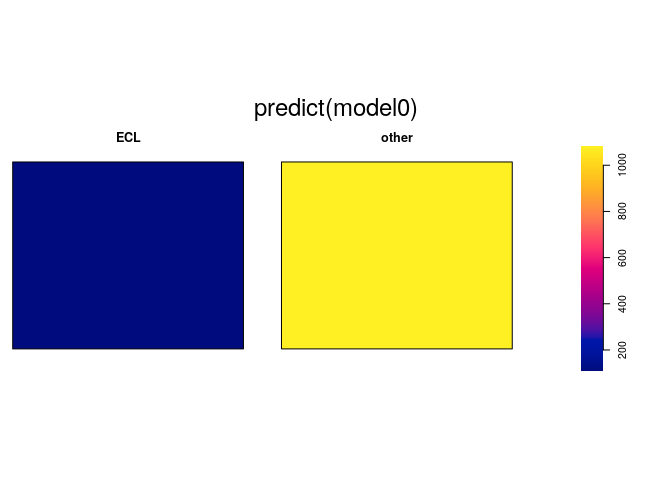
In the formula, the marks variable is a categorical variable. The
effect of the model formula mucosa ~ marks is to estimate a different
intensity for each level, that is, a different intensity for each type
of point. The model formula mucosa ~ marks is equivalent to saying
that the intensity of the points of type
is
for each where
are the different constant intensities
to be estimated. The actual printed output will depend on the convention
for handling “contrasts” in linear models.
The marks variable can be combined with other explanatory variables:
model1 <- ppm(mucosa ~ marks + y)
model1
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks + y
##
## Fitted trend coefficients:
## (Intercept) marksother y
## 5.131273 2.286730 -1.156055
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 5.131273 0.1164479 4.903039 5.3595066 *** 44.064966
## marksother 2.286730 0.1112542 2.068675 2.5047840 *** 20.554089
## y -1.156055 0.1406821 -1.431787 -0.8803236 *** -8.217504
coef(model1)
## (Intercept) marksother y
## 5.131273 2.286730 -1.156055
plot(predict(model1))
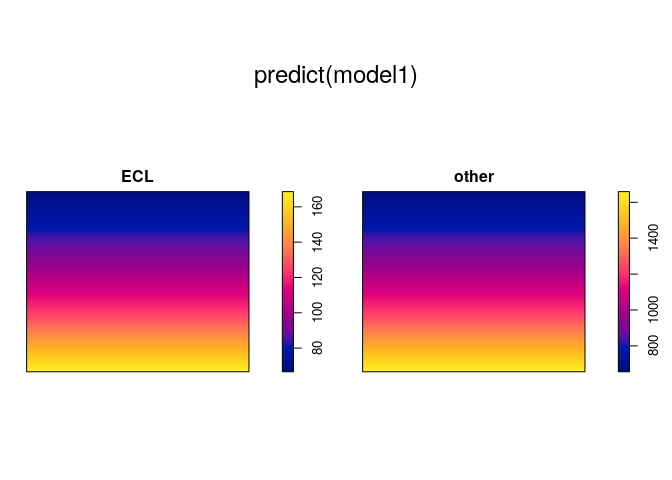
The model formula ~marks + y states that
where and
are
parameters. That is, the dependence on the
coordinate has the same
“slope” coefficient
for
each type of point, but different types of points have different
abundance overall.
## This requires spatstat 1.60-1.006 or later
if(packageVersion("spatstat") < "1.60-1.006"){
message("This version of spatstat cannot produce the relevant type of effect plot.")
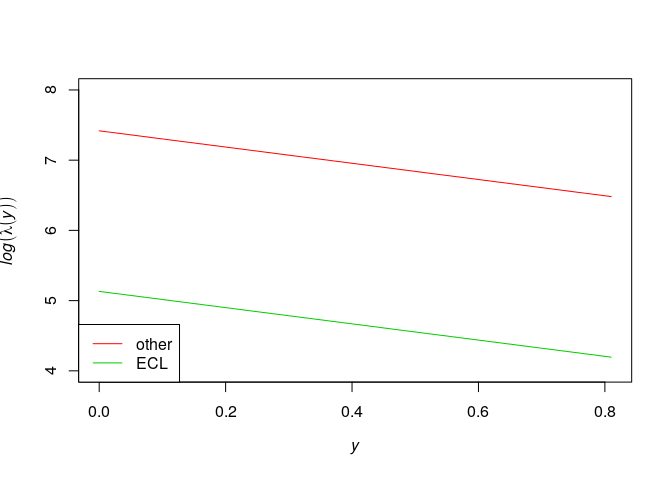
} else{
plot(effectfun(model1, "y", marks="other"),
log(.y) ~ .x, ylim=c(4,8), col=2, main="")
plot(effectfun(model1, "y", marks="ECL"),
add=TRUE, col=3, log(.y) ~ .x)
legend("bottomleft", lwd=c(1,1), col=c(2,3), legend=c("other", "ECL"))
}
model2 <- ppm(mucosa ~ marks * y)
model2
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks * y
##
## Fitted trend coefficients:
## (Intercept) marksother y marksother:y
## 5.884603 1.452251 -3.862202 2.938790
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 5.884603 0.1635842 5.563984 6.205222 *** 35.972936
## marksother 1.452251 0.1749458 1.109364 1.795139 *** 8.301149
## y -3.862202 0.5616953 -4.963104 -2.761299 *** -6.875973
## marksother:y 2.938790 0.5804890 1.801053 4.076528 *** 5.062611
coef(model2)
## (Intercept) marksother y marksother:y
## 5.884603 1.452251 -3.862202 2.938790
plot(predict(model2))
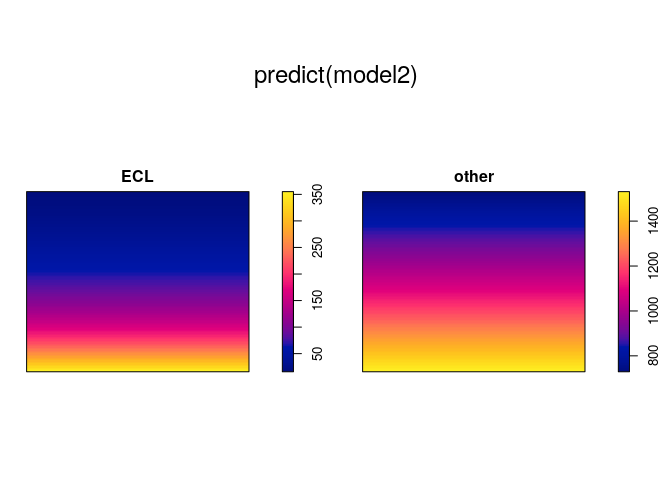
The model formula ~marks * y states that
where and
are parameters. The intensity may depend
on the
coordinate in a
completely different way for different types of points.
## This requires spatstat 1.60-1.006 or later
if(packageVersion("spatstat") < "1.60-1.006"){
message("This version of spatstat cannot produce the relevant type of effect plot.")
} else{
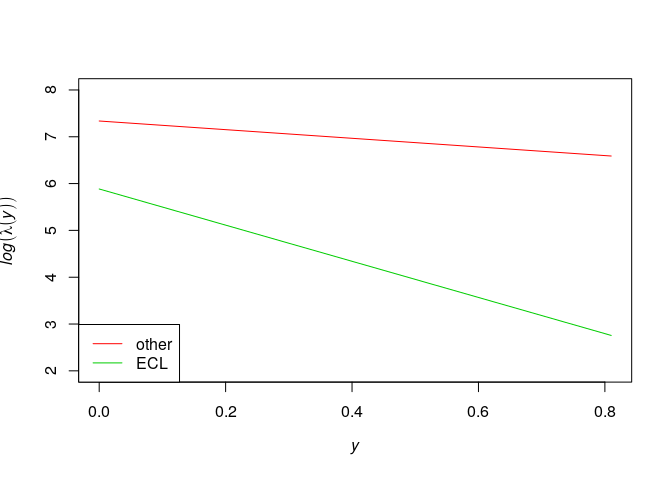
plot(effectfun(model2, "y", marks="other"),
log(.y) ~ .x, col=2, ylim=c(2,8), main="")
plot(effectfun(model2, "y", marks="ECL"),
add=TRUE, col=3, log(.y) ~ .x)
legend("bottomleft", lwd=c(1,1), col=c(2,3), legend=c("other", "ECL"))
}
Other examples to discuss:
model1xy <- ppm(mucosa ~ marks + x + y)
model1xy
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks + x + y
##
## Fitted trend coefficients:
## (Intercept) marksother x y
## 5.135026706 2.286729721 -0.007512035 -1.156055806
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 5.135026706 0.1290806 4.8820333 5.3880201 *** 39.7815429
## marksother 2.286729721 0.1112542 2.0686754 2.5047840 *** 20.5540892
## x -0.007512035 0.1115200 -0.2260873 0.2110632 -0.0673604
## y -1.156055806 0.1406820 -1.4317876 -0.8803241 *** -8.2175076
coef(model1xy)
## (Intercept) marksother x y
## 5.135026706 2.286729721 -0.007512035 -1.156055806
plot(predict(model1xy))
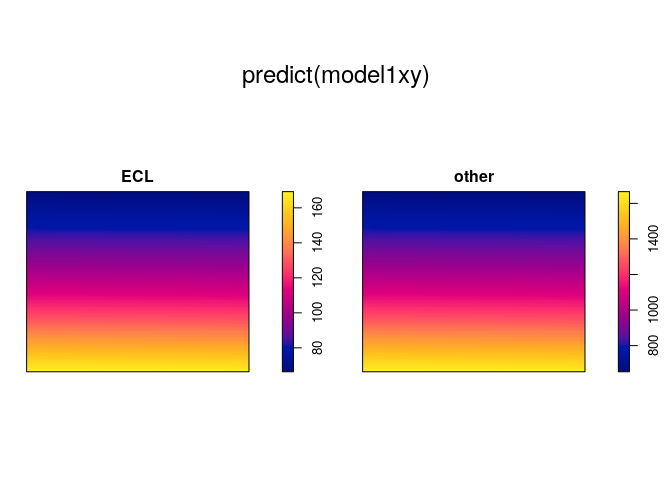
model2xy <- ppm(mucosa ~ marks * (x + y))
model2xy
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks * (x + y)
##
## Fitted trend coefficients:
## (Intercept) marksother x y marksother:x
## 5.85306325 1.49110635 0.06275125 -3.86220183 -0.07739871
## marksother:y
## 2.93878953
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 5.85306325 0.2473601 5.3682464 6.3378801 *** 23.6621186
## marksother 1.49110635 0.2616142 0.9783520 2.0038607 *** 5.6996392
## x 0.06275125 0.3672459 -0.6570374 0.7825399 0.1708699
## y -3.86220183 0.5616962 -4.9631062 -2.7612975 *** -6.8759620
## marksother:x -0.07739871 0.3854477 -0.8328623 0.6780649 -0.2008021
## marksother:y 2.93878953 0.5804899 1.8010502 4.0765289 *** 5.0626022
coef(model2xy)
## (Intercept) marksother x y marksother:x
## 5.85306325 1.49110635 0.06275125 -3.86220183 -0.07739871
## marksother:y
## 2.93878953
plot(predict(model2xy))
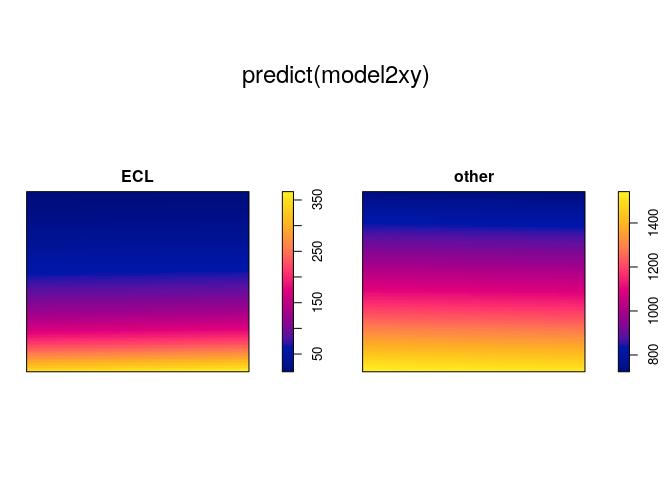
model3 <- ppm(mucosa ~ marks + polynom(x, y, 2))
model3
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks + (x + y + I(x^2) + I(x * y) + I(y^2))
##
## Fitted trend coefficients:
## (Intercept) marksother x y I(x^2) I(x * y)
## 4.8407551 2.2867297 0.2828123 0.8046386 -0.2449076 -0.1328756
## I(y^2)
## -2.5066916
##
## Estimate S.E. CI95.lo CI95.hi Ztest Zval
## (Intercept) 4.8407551 0.1865057 4.4752107 5.2062995 *** 25.9550007
## marksother 2.2867297 0.1112542 2.0686754 2.5047840 *** 20.5540892
## x 0.2828123 0.4840067 -0.6658233 1.2314479 0.5843149
## y 0.8046386 0.6087936 -0.3885748 1.9978521 1.3216937
## I(x^2) -0.2449076 0.4346487 -1.0968034 0.6069882 -0.5634611
## I(x * y) -0.1328756 0.5195314 -1.1511385 0.8853873 -0.2557605
## I(y^2) -2.5066916 0.7025926 -3.8837479 -1.1296354 *** -3.5677739
coef(model3)
## (Intercept) marksother x y I(x^2) I(x * y)
## 4.8407551 2.2867297 0.2828123 0.8046386 -0.2449076 -0.1328756
## I(y^2)
## -2.5066916
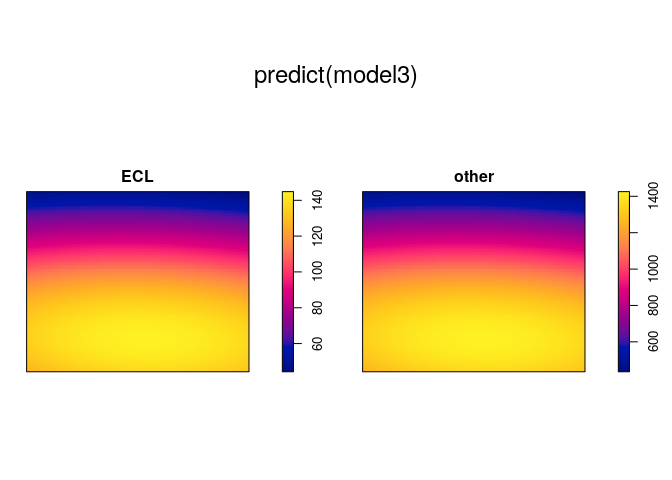
plot(predict(model3))
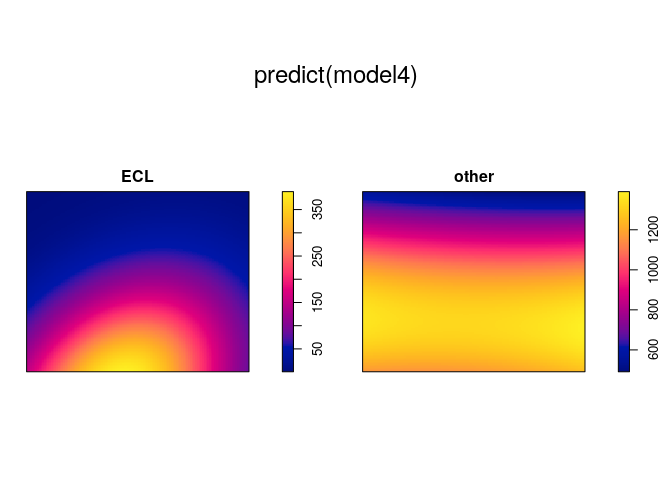
model4 <- ppm(mucosa ~ marks * polynom(x,y,2))
model4
## Nonstationary multitype Poisson process
##
## Possible marks: 'ECL' and 'other'
##
## Log intensity: ~marks * (x + y + I(x^2) + I(x * y) + I(y^2))
##
## Fitted trend coefficients:
## (Intercept) marksother x
## 5.018632 2.040831 4.310737
## y I(x^2) I(x * y)
## -2.261047 -4.905847 3.086670
## I(y^2) marksother:x marksother:y
## -5.199005 -4.388268 3.579736
## marksother:I(x^2) marksother:I(x * y) marksother:I(y^2)
## 5.059764 -3.342710 2.439852
##
## Estimate S.E. CI95.lo CI95.hi Ztest
## (Intercept) 5.018632 0.5006414 4.0373924 5.9998706 ***
## marksother 2.040831 0.5278293 1.0063043 3.0753570 ***
## x 4.310737 1.7516879 0.8774918 7.7439820 *
## y -2.261047 2.2251226 -6.6222072 2.1001133
## I(x^2) -4.905847 1.6650184 -8.1692227 -1.6424705 **
## I(x * y) 3.086670 2.6191912 -2.0468503 8.2201905
## I(y^2) -5.199005 3.0833891 -11.2423363 0.8443271
## marksother:x -4.388268 1.8233568 -7.9619815 -0.8145540 *
## marksother:y 3.579736 2.3165955 -0.9607074 8.1201802
## marksother:I(x^2) 5.059764 1.7252514 1.6783335 8.4411947 **
## marksother:I(x * y) -3.342710 2.6736718 -8.5830101 1.8975907
## marksother:I(y^2) 2.439852 3.1699025 -3.7730426 8.6527470
## Zval
## (Intercept) 10.0244036
## marksother 3.8664599
## x 2.4609047
## y -1.0161449
## I(x^2) -2.9464219
## I(x * y) 1.1784822
## I(y^2) -1.6861331
## marksother:x -2.4066971
## marksother:y 1.5452574
## marksother:I(x^2) 2.9327692
## marksother:I(x * y) -1.2502319
## marksother:I(y^2) 0.7696931
coef(model4)
## (Intercept) marksother x
## 5.018632 2.040831 4.310737
## y I(x^2) I(x * y)
## -2.261047 -4.905847 3.086670
## I(y^2) marksother:x marksother:y
## -5.199005 -4.388268 3.579736
## marksother:I(x^2) marksother:I(x * y) marksother:I(y^2)
## 5.059764 -3.342710 2.439852
plot(predict(model4))
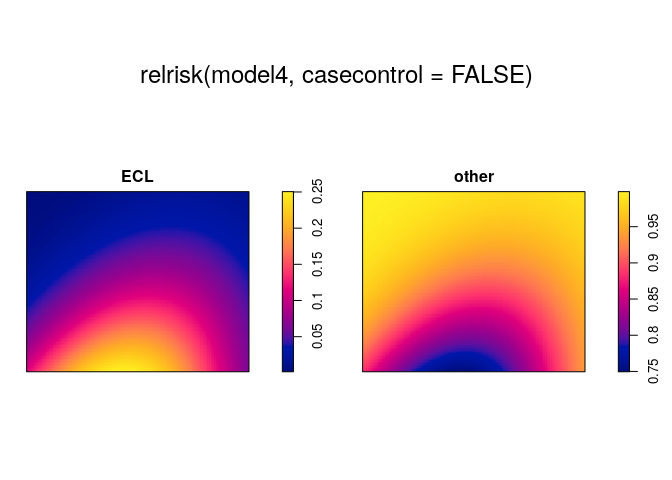
Parametric estimation of spatially-varying probability
When we have fitted a point process model to a multi-type point pattern, we can compute ratios of the intensities of different types. This is automated in relrisk.ppm:
plot(relrisk(model4, casecontrol=FALSE))
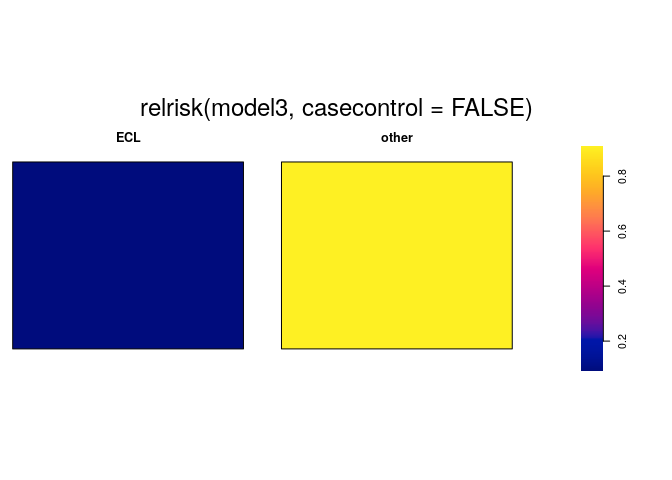
plot(relrisk(model3, casecontrol=FALSE), equal.ribbon=TRUE)
Test for segregation
One way to test for segregation is to compare two models, with the null model stating that there is no segregation:
nullmodel <- ppm(mucosa ~ marks + polynom(x, y, 2))
altmodel <- ppm(mucosa ~ marks * polynom(x, y, 2))
anova(nullmodel, altmodel, test="Chi")
## Analysis of Deviance Table
##
## Model 1: ~marks + (x + y + I(x^2) + I(x * y) + I(y^2)) Poisson
## Model 2: ~marks * (x + y + I(x^2) + I(x * y) + I(y^2)) Poisson
## Npar Df Deviance Pr(>Chi)
## 1 7
## 2 12 5 44.834 1.568e-08 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1